Due to Aspera license limitations, users have to install the software under their home to user the software.
For example, for user abc123, the working directory will be
srun --pty -p interactive -t 0-12:0:0 --mem 2000MB -n 1 /bin/bash
mkdir /n/scratch/users/${USER:0:1}/${USER}/testDbGaP
cd /n/scratch/users/${USER:0:1}/${USER}/testDbGaP
module load sratoolkit/2.10.7 |
# Configure sratoolkit
vdb-config --interactive
# Directly press x key to quit
# By default, sratoolkit uses working diretory as cache. It is better to use scratch instead:
echo /repository/user/main/public/root = \"/n/scratch/users/${USER:0:1}/${USER}/ncbi\" >> ~/.ncbi/user-settings.mkfg
|
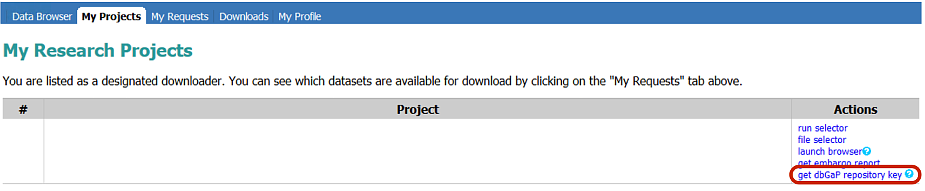
# Upload the dbGaP repository Key to O2: scp ~/Download/prj_phs710EA_test.ngc $USER@transfer.rc.hms.harvard.edu:~/.ncbi |
# Load sratookit module module load sratoolkit/2.10.7 # Use prefetch to download SRA file. prefetch --ngc ~/.ncbi/prj_phs710EA_test.ngc -p SRR1219902 # Convert SRA file to FASTQ with fastq-dump. -X 5 means only gives first five reads to give a quick test. fastq-dump -X 5 --ngc ~/.ncbi/prj_phs710EA_test.ngc --split-files SRR1219902 |