| Table of Contents | ||||
|---|---|---|---|---|
|
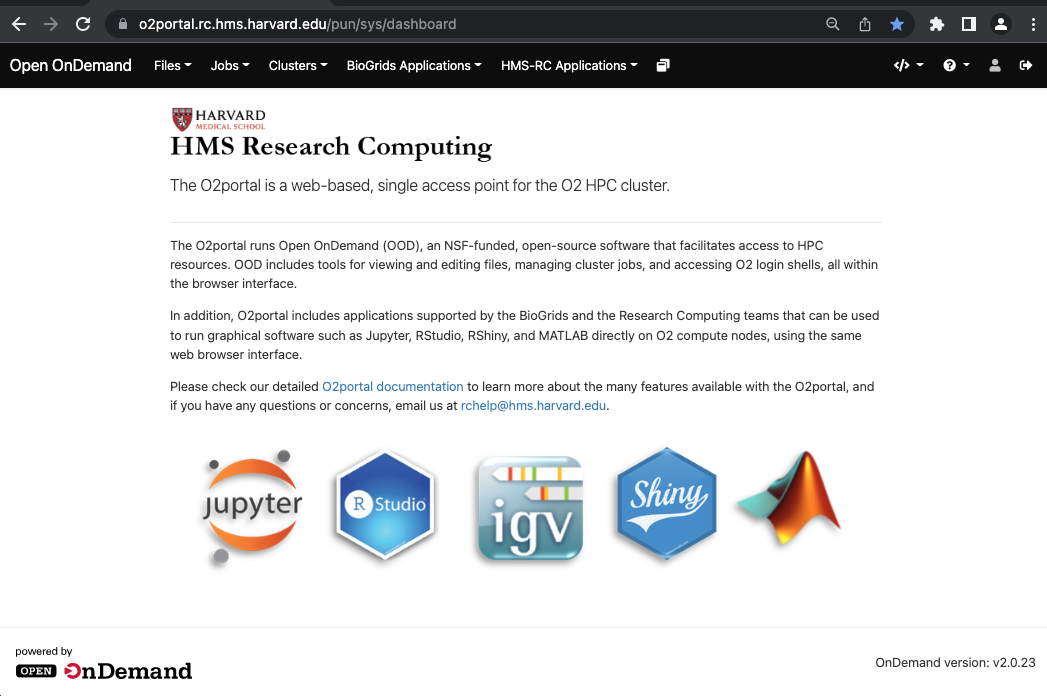
The O2Portal runs Open OnDemand (OOD), an NSF-funded, open-source software that facilitates access to HPC resources. Open OnDemand was created and is maintained by the Ohio Supercomputer Center.
The O2Portal provides easy access to HMS’ O2 cluster and includes tools for viewing/editing files, managing cluster jobs, accessing login shells and more, entirely via the same single-sign-on web-based interface.
How to login to O2Portal
| Note |
|---|
You must have an O2 account in order to use the O2Portal |
Navigate on your web browser to https://o2portal.rc.hms.harvard.edu and authenticate using your HMS ID and password.
After entering the HMS ID and Password, you will be prompted to complete the two-factor authentication step (2FA) to finally land on the main O2Portal page:
| Warning |
|---|
Some of Open OnDemand’s features and apps might not work properly when using the Safari web browser. We recommend using Google Chrome with O2Portal. |
Features available in O2Portal
Open OnDemand includes several useful tools that can be accessed from your web browser; please use the links below to learn more details about each O2Portal tool.
Computational Applications available in O2Portal
It is possible to run a number of computational applications such as Jupyter, RStudio, MATLAB and more as O2 cluster jobs directly from within the O2Portal website.
Each O2Portal application, once launched, will submit an actual job on the O2 cluster and the application will become available once the job is dispatched by the Slurm scheduler.
This feature allows you to run an interactive graphical application as if it’s running locally on your desktop, but using O2 compute nodes with large RAM (memory), multiple CPU cores and GPU availability.
Applications will not stop running if the web browser connection to the O2Portal is interrupted and will continue running until the O2 jobs run out of their allocated wall-time or the user deletes the application instance.
This feature allows any user to run long interactive applications which remains accessible at a later time and even from a different computers. You can start your O2Portal Application from your work computer and reconnect to the same application from your home machine.
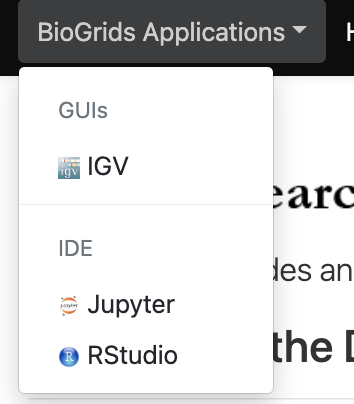
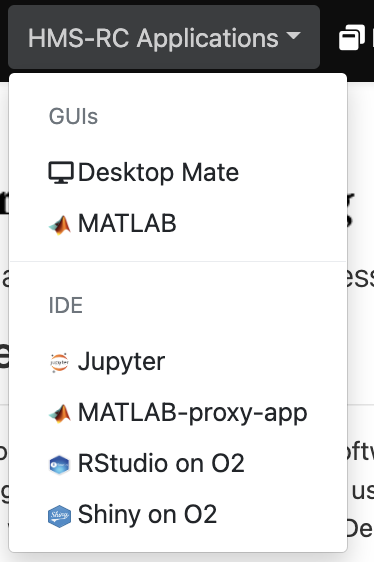
The BioGrids and the HMS Research Computing teams provide two sets of scientific software applications for the O2portal. The BioGrids team pre-configures its applications to ensure that they will run compatibly with each other. HMS Research Computing applications are more open to user configuration. We invite you to use whichever tools best meet your needs.
To select an application click on the desired O2Portal tab application from the BioGrids or the HMS-RC drop menus
To see your running interactive applications click on the My Interactive Sessions tab
If you don’t have any running application you should see a page like:
Please check the links below for specific information on the available O2Portal apps.
HMS Research Computing Applications
BioGrids Applications
| Note |
|---|
When using the applications described you will be submitting and running jobs on the O2 cluster. Please make sure to read our O2 documentation to familiarize yourself with how the O2 cluster and the Slurm scheduler work. |
| Note |
|---|
Note that if your Lab is paying for using the O2 cluster resources any O2 jobs dispatched using the O2Portal, including interactive applications, will also be included in your quarterly bill. |
F.A.Q.
Please check our O2portal FAQ link for the most commonly asked questions about O2Portal and OOD./